Machine Learning Enabled Materials Production
The core capability to produce precise and complex biomolecular transformations in cell lysates allows for the construction of enzyme libraries to create large datasets enabling the use of machine learning methods to produce novel chemicals and materials. Whereas it may take months to test unique bioproduction pathways, using cell-free bioprototyping my lab will be able to rapidly generate enzymatic libraries that can be combinatorially reconstituted with even larger substrate libraries into full metabolic pathways in a fraction of the time.
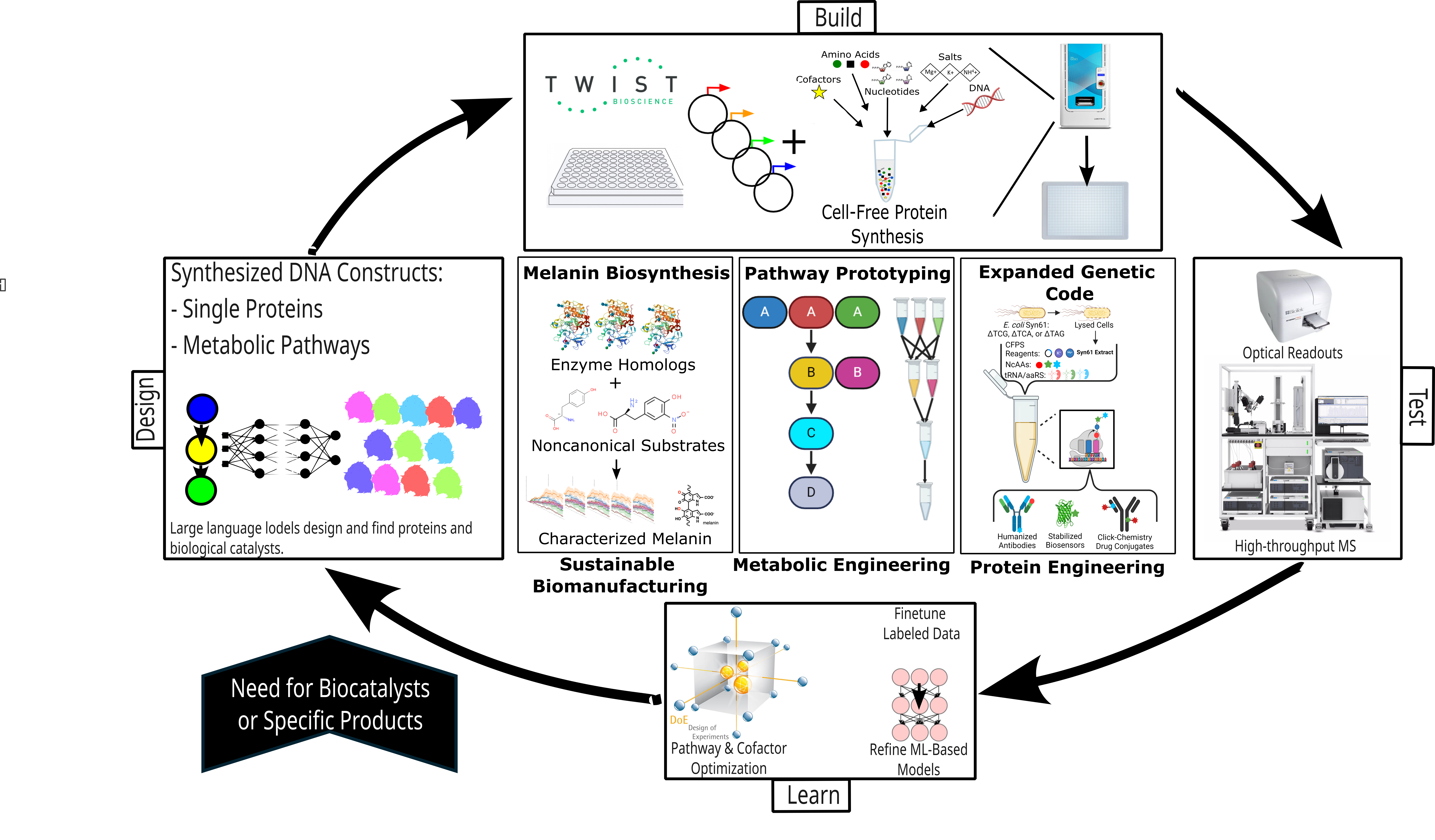
One of our first targets, melanin, a natural biopolymer known for its unique ability to absorb a wide range of radiations is a versatile material for coatings, absorbents, and even as a building material. However, the mechanisms by which melanins are synthesized are often environmentally unfriendly, expensive chemical synthesis, or inefficient due to cellular toxicity. Beyond simply producing large amounts of melanin, my goal for this work is two-fold. First, explore the use of substrates like noncanonical amino acids and multipeptides as sources for novel melanins with improved properties like heat resistance, magnetism, and color variations; and second, to fuse experimental activity data with ligand docking, and physicochemical properties of proteins and ligands with various machine learning models into accurate predictions of substrate scope.
Immediate Work: Train LLM-based models to test novel substrates and biocatalysts for biological polymer production.
Long term Vision: Enable rapid and automated DBTL loops to effectively engineer enzymes, pathways, and synthetic biology tools.