Portfolio item number 1
Short description of portfolio item number 1
Short description of portfolio item number 1
Short description of portfolio item number 2 
Garcia DC, Xiaolin C, Land M, Standaert RF, Morrell-Falvey JL, Doktycz MJ. ACS Chemical Biology, 2019.
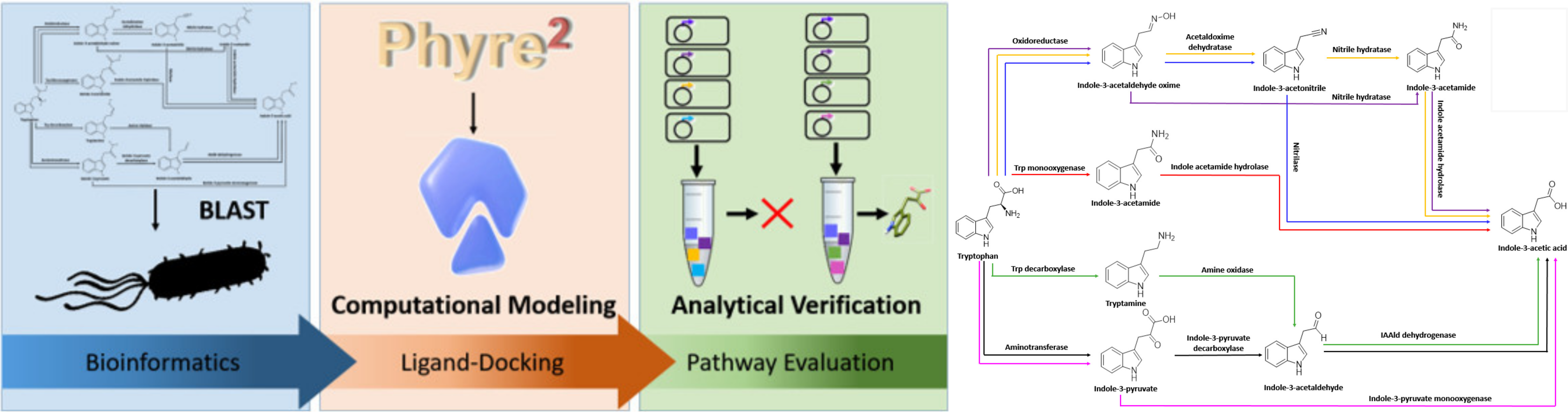
Elucidating the interaction networks associated with secondary metabolite production in microorganisms is an ongoing challenge made all the more daunting by the rate at which DNA sequencing technology reveals new genes and potential pathways. Developing the culturing methods, expression conditions, and genetic systems needed for validating pathways in newly discovered microorganisms is often not possible. In this work we describe an in vitro computationally assisted pathway description approach that employs bioinformatic searches of genome databases, protein structural modeling, and protein–ligand-docking simulations to predict the gene products most likely to be involved in a particular secondary metabolite production pathway. This information is then used to direct in vitro reconstructions of the pathway and the combined computational culling and in vitro verification approach used to define the functional capabilities of the component enzymes and the tryptophan to IAA pathway in Pantoea sp. YR343.
[Link] [pdf]Garcia DC , Dinglasan E , Shrestha H, Abraham PE, Hettich RL, Doktycz MJ. Met Eng Comms, 2021.

Cell-free systems harness the metabolic potential of diverse organisms in an open and modifiable environment. Removing the cellular context provides the ability to produce biological products without the need to maintain cell viability and enables metabolic engineers to explore novel chemical transformation systems. However, only limited tools are available for engineering the contents of the extracts used for cell-free systems. While environmental variables of a cell-free system can be easily manipulated, the proteomic content of the crude extract is more difficult to engineer. We describe a framework to enable the removal of specific proteins from crude extracts for CFME resulting in a modified proteome capable of producing engineered metabolic phenotypes not possible in a living cells with minimal impact on the viability of the donor cell.
[Link] [pdf]Garcia DC , Davies JP , Rhea K, Lee MF, Lux MM. bioRxiv, 2023.

Porphyrins are molecules composed of one or more cyclic tetrapyrroles whose aromaticity enable semiconductor-like properties, making them useful in a broad range of applications, including artificial photosynthesis and light harvesting, catalysis, single-molecule electronics, sensors, nonlinear optics, and chemical warfare agent degradation. Due to its utility in this work we take advantage of porphyrins as both molecules of interest and as an easily detectable products in a cell-free extract to i) show our ability to produce porphyrins using enriched cell-free extracts, ii) explore consolidating the extract source cells into a single co-culture fermentation in order to limit the need for multiple extract productions, and iii) rapidly generate ideal cofactor and substrate mixtures using DBTL-cycles powered by Design of Experiments (DOE).
[Link] [pdf]Garcia DC , Davies JP , Davidson CE, Philips DA, Miklos AE, Lux MW. bioRxiv, 2024.
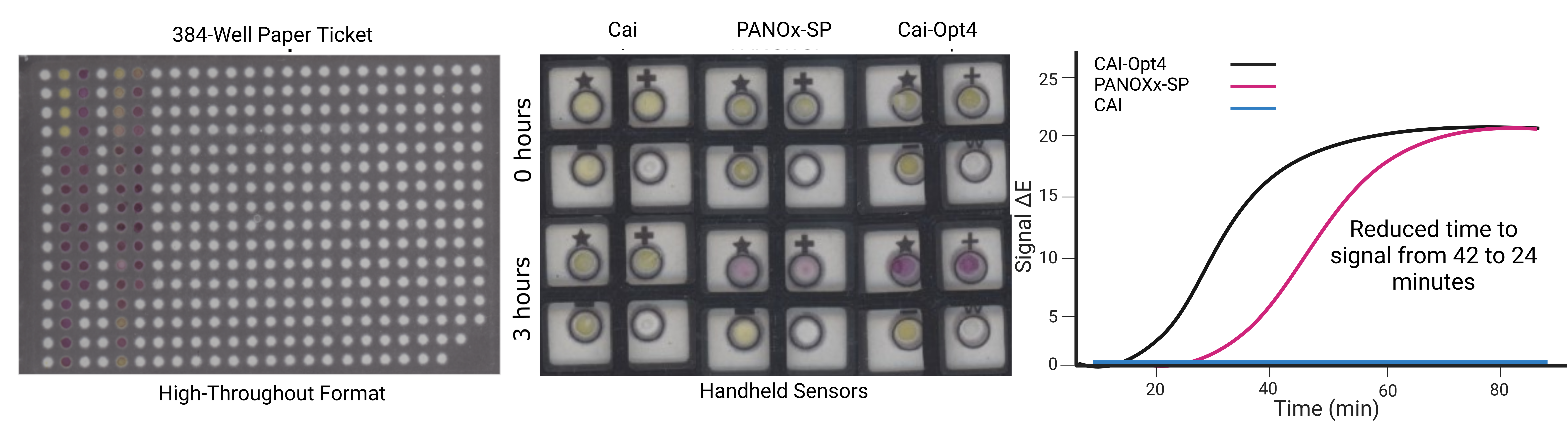
Cell-free expression systems maintain core cellular processes without intact cells and offer attractive properties as point-of-need biosensors. The ability to lyophilize, store, and use on demand makes these sensors usable in the field, and the lack of membranes means there are no analyte transport issues and new sensors can be deployed by simply adding a different DNA molecule. We present new high-throughput screening methods to optimize cell-free expression formulations when embedded into paper for use as sensors. Our method leverages acoustic liquid handling to dispense reactions onto 384-well paper ticket formats and machine vision to quantify reaction performance from a colorimetric reporter enzyme. The throughput enabled shifts the bottleneck from experimental execution to selecting the experiments We showcase the use of the cost-optimized formulation in a 3D-printed paperfluidic device where it outperforms the standard formulation at much lower cost.
[Link] [pdf]Undergraduate course, University 1, Department, 2014
This is a description of a teaching experience. You can use markdown like any other post.
Workshop, University 1, Department, 2015
This is a description of a teaching experience. You can use markdown like any other post.